‡ Joint first authors
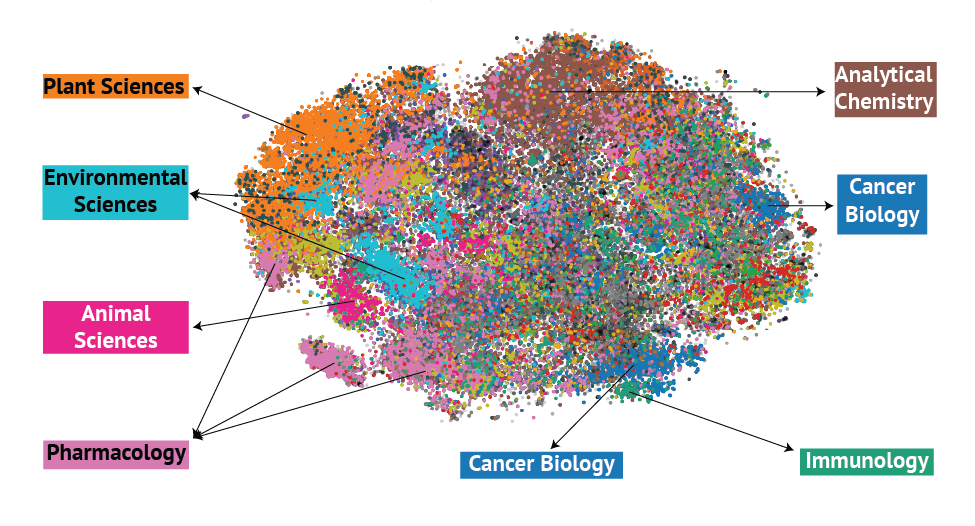
A Large Language Model-Powered Map of Metabolomics Research
Analytical Chemistry, 2025
A comprehensive analysis of metabolomics research trends using large language models to map the evolution and future directions of the field.
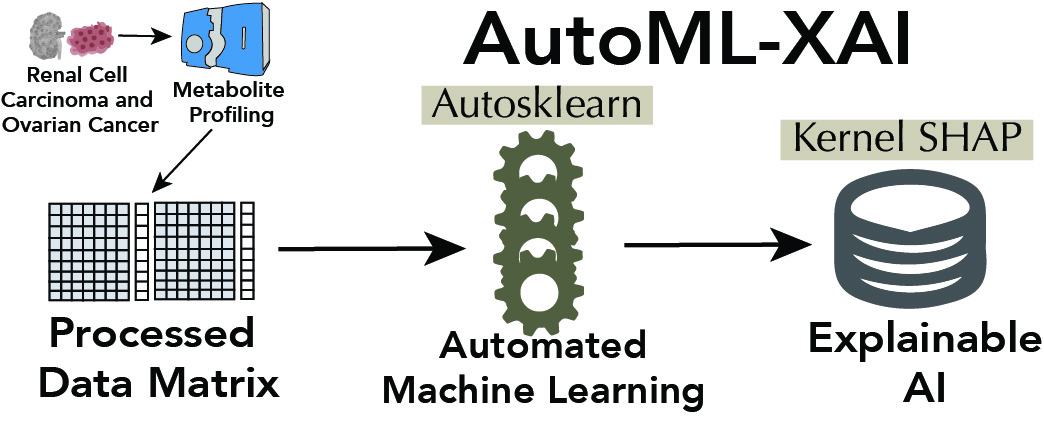
Automated machine learning and explainable AI (AutoML-XAI) for metabolomics: improving cancer diagnostics
Journal of the American Society for Mass Spectrometry, 2024
Development of automated machine learning pipelines with explainable AI for enhancing cancer biomarker discovery in metabolomics datasets.
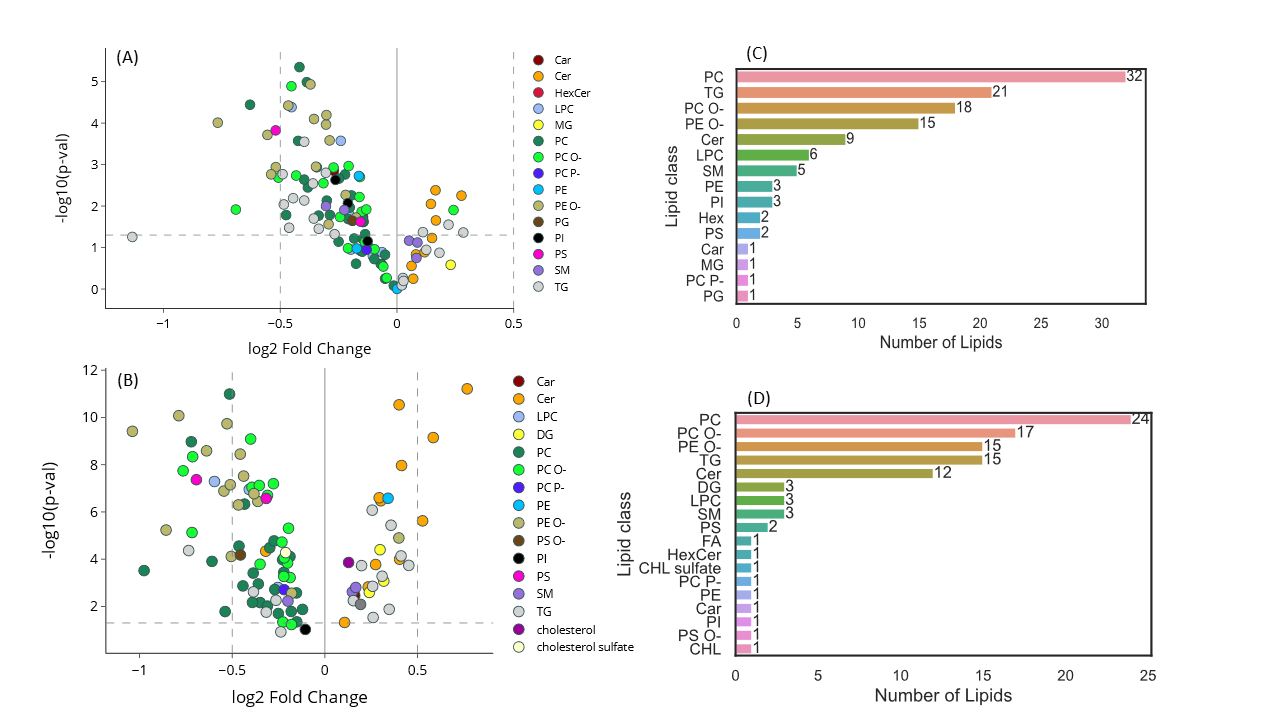
Serum Lipidome Profiling Reveals a Distinct Signature of Ovarian Cancer in Korean Women
Provisional patent was filed for the findings from this work.
Identification of serum lipid biomarkers for ovarian cancer detection using advanced metabolomics and machine learning approaches.
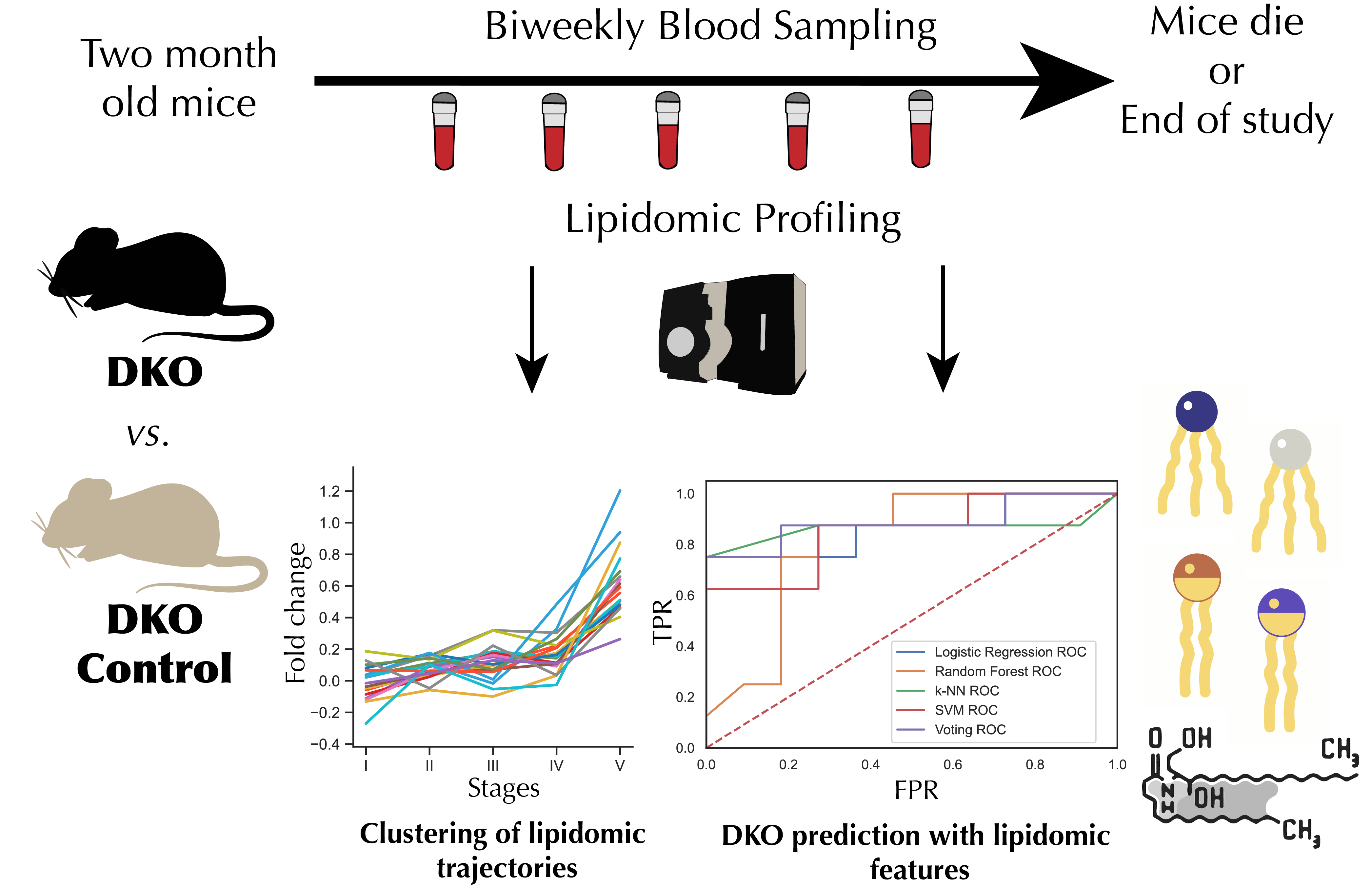
Machine Learning Reveals Lipidome Remodeling Dynamics in a Mouse Model of Ovarian Cancer
Journal of Proteome Research, 2023
Comprehensive analysis of lipid metabolism changes during ovarian cancer progression using machine learning and multi-platform metabolomics.
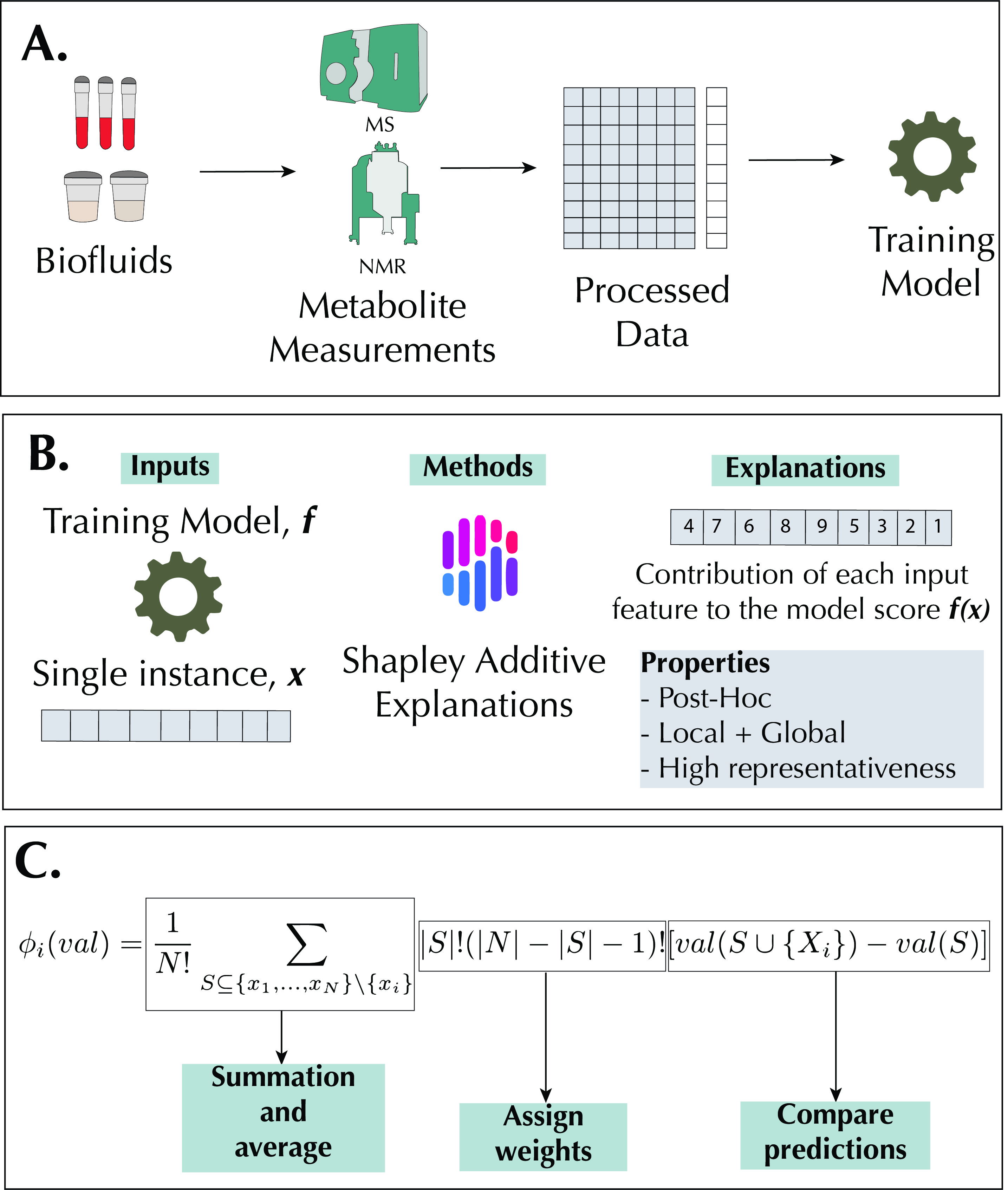
Interpretable machine learning with tree-based shapley additive explanations: Application to metabolomics datasets for binary classification
Development and validation of interpretable machine learning methods for metabolomics data analysis with focus on clinical applications.
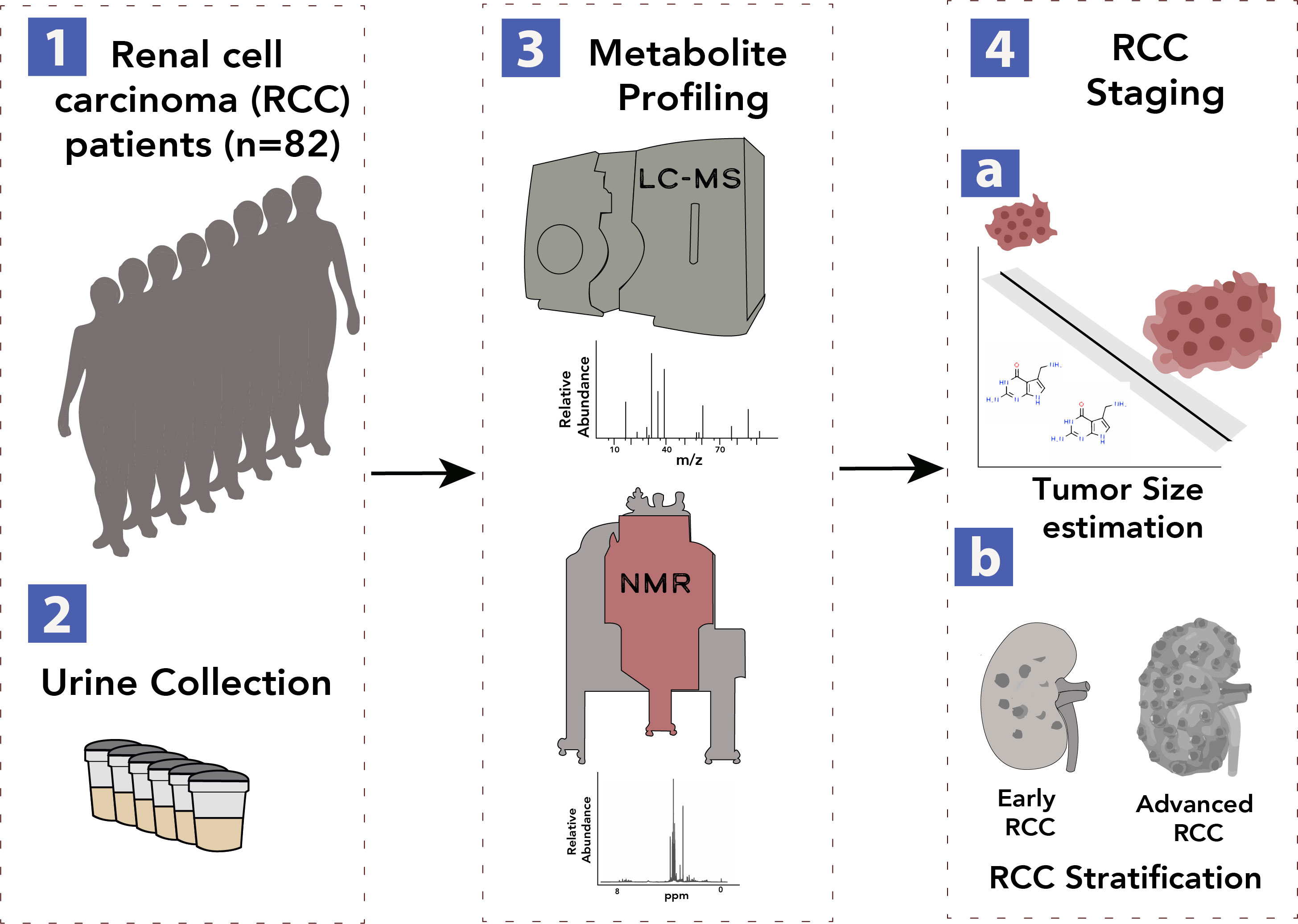
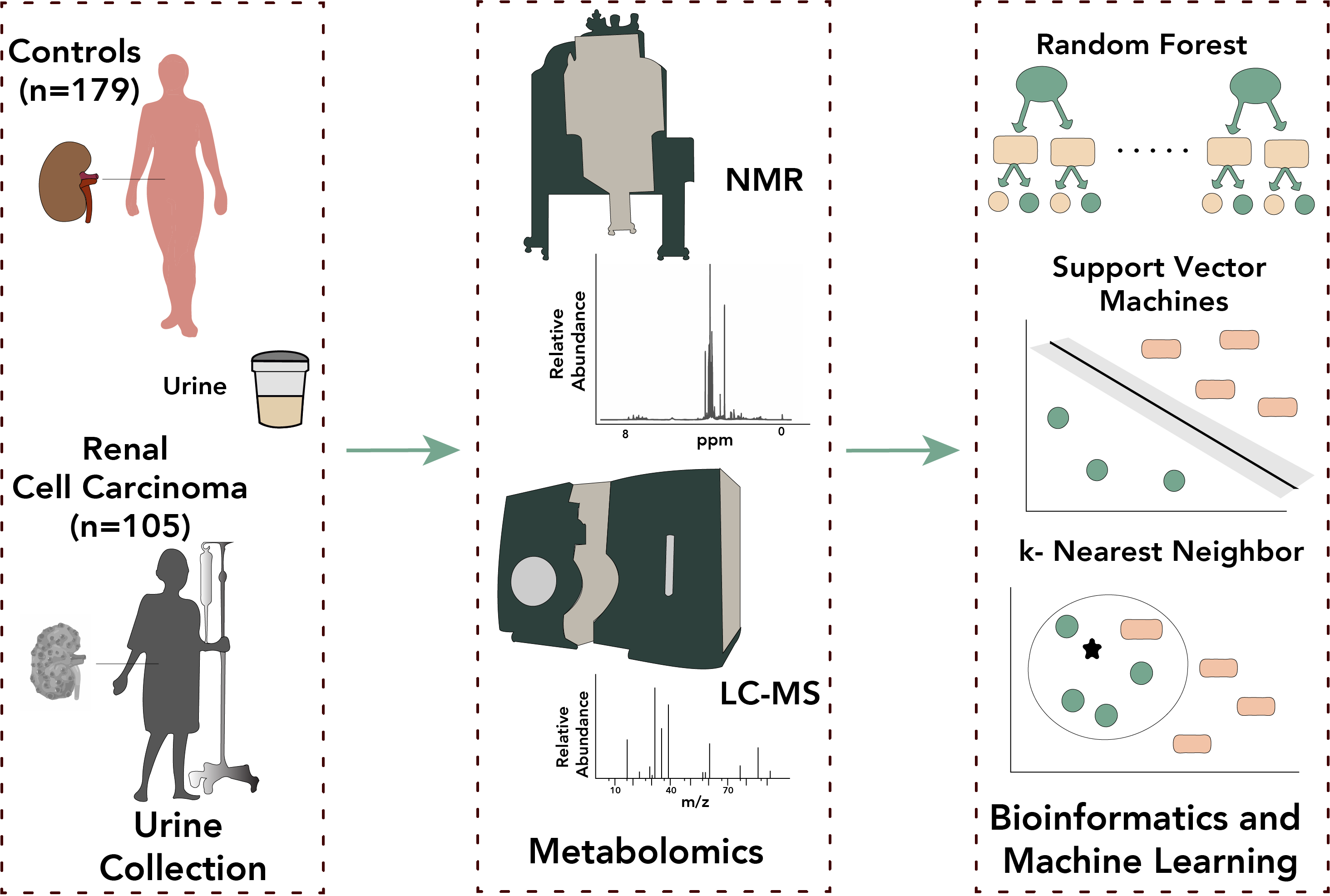
Urine-Based Metabolomics, and Machine Learning Reveals Metabolites Associated with Renal Cell Carcinoma Stage
Investigation of urine metabolites associated with different stages of renal cell carcinoma using machine learning approaches.
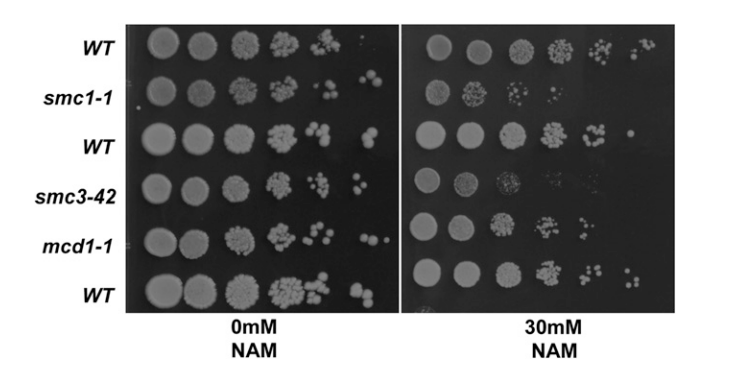
A Genome-Wide Screen with Nicotinamide to Identify Sirtuin-Dependent Pathways in Saccharomyces cerevisiae
Systematic investigation of sirtuin-dependent pathways in yeast using genome-wide screening approaches.